A Walk Through Parameter Space
This is just for me and my note taking as it is for anyone else. I'm trying to take notes and come up with a plan and actually make progress on a project all in real time. Broad strokes goal of this project is to recreate electrophysiology properties from morphology and transcriptome data. No prizes for guessing why I want to do this, especially because it was laid out in the last post. the tl;dr is that the Allen institute has a lot of data on morphology, electrophysiology, and transcriptome (MET) but they don't have a lot of neuron models fit to that data. So I kind of have to make my own neuron model fit to these parameters. You might naively think this is easy. By you, I of course mean me. Anyway, the Allen Institute did publish a paper on this which is cool and I am now trying to copy. I made my own neuron model first based on this kind of outdated paper but the principle is all the same. The following are my notes on this process.
So if you take the "obvious" first pass of just trying to run a genetic algorithm on the MSE of the membrane potential just given a certain current stimulus, you get crappy results. What I am trying, and they suggest in the paper, is to fit progressively. So first get things like the resting potential and the time constants for non spiking activity fit and then move on to tuning channel conductances and what not. It remain to be seen by a guy like me if this works. First step is resting potential. The neuron model I copied has a resting potential of ~-50mV which is not what most of the Allen institute data has. What I did was vary the capacitance, membrane area, conductances, and leak reverse potential then found pearson r values correlating those with the resting potential. Turns out, only 3 of these have a statistically significant correlation, those are the leak conductance (not surprising), H conductance (it's a kind of catch all ion in the model, I don't care enough about the actual ground truth biophysics at this stage to explain what this is), and then the membrane area. Oh the Leak and H are really pulling their weight, area is barely significant so I think I'm just going to ignore it in everything.
Next we want to get magnitude and time constants for non spiking changes. What exactly does that mean? If you inject the cell with some current in a square wave, but not enough current to cause it to spike, it will approach some new "resting" state generally in an exponential fashion. Sometimes it will kind of overshoot and then decay back down, this is the start of a spike. Sometimes the decay will take so long it won't really be close to a new "resting" state by the time the square wave stimulus repeats, this is just a function of the frequency of the square wave.
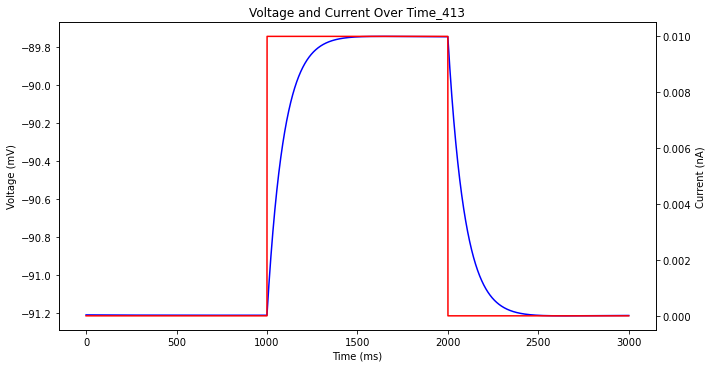
This is what most of the data from the allen institute looks like. Unfortunately, most of mine does not look like that which is quite frustrating I must admit. So after some testing I have come to the conclusion that H and Leak conductances are the most important parameter for this general shape as well. If H is very low and Leak is very high then you will almost always get shapes like this which is epic. God, this search space is enormous. Ok so I am going to track a few things, just kind of stream of consciousness thing
- time of first spike
- current during first spike
- peak voltage
- resting potential
- minimum voltage
- time constants and delta V for let's say -0.1, -0.05, 0.05, and 0.1 nA
and I'm going to do that by taking steps through
- Leak reversal potential
- H conductance
- Leak conductance
with the rest of the parameters being randomized.
I can have at least 5000 neurons simulated simultaneously.
This push has the ranges I am going to use, I mean they aren't formalized but it has a good mix of spiking behavior and the decay properties.
This is pretty arbitrary but I think I am going to have a range of 8 values for each of H, Leak, and leak reversal and simulate 1000 neurons with randomized values for the other parameters. This gives ~half a million template neurons to feed into the Allen Institute data.
Hey! I did that thing I described.
It took a while but it is all run now. What is unfortunate is that the resting potential of the neuron seems to be grossly inconsistent. One some it's like -62, some it's -78 mV. Maybe I'm the dummy that has grouped the cells wrong but honestly I don't think so. I don't know what the best way to proceed is. I guess only use recordings from one session? That does give pretty consistent results but what will this say for the later morphology + transcriptome -> ephys feature prediction? How can I predict ephys if that is not consistent? A smart man would look at categorize the stimulus patterns and look at differences in the derived electrophysiology properties to see if variance between cells is greater than variance within cells but hey, that sounds like a lot of work.
It's a new day folks. Quite frankly I feel like I just put my brain in a woodchipper trying to debug. I am ~85% sure my code is right so there's probably a 15% chance it actually is. Here is the updated version of the neuron simulation with hopefully the final units. Also that has the nwb data loader. Bottom line is this, I have no real justification past vibes but I am going to multiply the current by 10x because it just makes sense, 0.05nA is too low to do anything with 99% of the time.
I think that 10x factor was a load of baloney! I took it away and tried fitting to only one current stimulus which still failed. I can't get the ramp or even a simple square wave to work. I mean they're close but miss some important features. Luckily I found this paper (actually I found it a while ago but I'm telling a story here, walk with me) that is exactly what I'm doing but better and they cite this other paper (that I also found before and foolishly ignored) and I'm hoping the extra ion channels they model has everything work out fine and dandy. Wow I just realized I had that Allen Paper in the intro, I guess I'm more tired than I thought. Say la vee as the french in france would say.
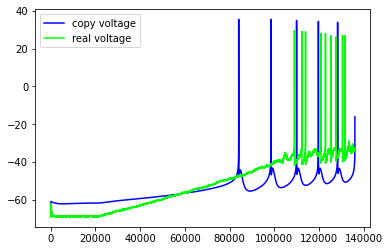
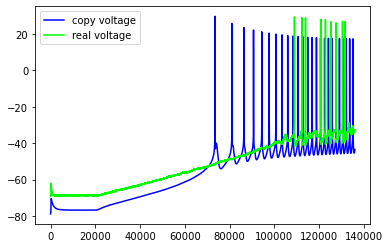
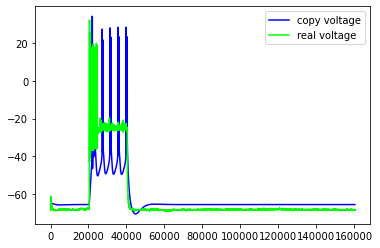
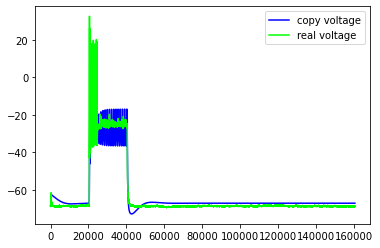
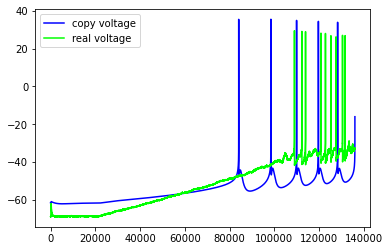
Just some examples of the model failing to capture some pretty important features of the real neuron
Hey after a lot of work, which I'm still not 100% done with, I believe I have made great progress implementing that new neuron model. Here is the code. There is something wrong with 2 of the ion channels, you get bad NaNs when they aren't 0. Also, the spikes are super low voltage, IDK what is causing that at all. Still here is an example of something pretty cool.
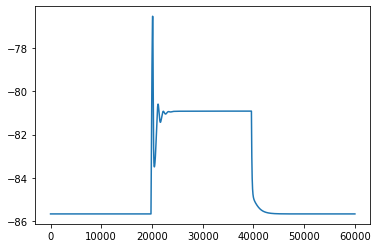
It does pretty crappy on the ramp, back to the old grindstone I guess.
I've tried a lot of stuff and it is all pretty frustrating. I am writing this 04/19/2025 and I will probably come back to this but maybe not. I have a few options:
- Debug in the hopes that I don't actually need the explicit axon part of this neuron model (seems likely based on all the other point neuron models I've worked on but annoying)
- Use NEURON
- Try and model the neurons as some kind of RNN with an input being the morphology and transcriptome data in addition to the current injection
- Ignore all the neuron modeling and just predict derived parameters (time of first spike, max frequency, peak action potential voltage, etc.)
I mean the goal is to see if electrophysiology is predictable based on morphology and transcriptomic data, no reason to kneecap myself working on making a neuron model that already exists and I'm too lazy to use right? Anyway, I'm going to try 2 and then 4 I guess? My main thing should actually be getting the MET data for all the neurons that is available from.
Update: because of my work on MET stuff I think there is a bug in this code about selecting single cells. I was trying to fit parameters of one model to multiple unrelated cells which obviously a no-go. Now, my model still lacks any morphology data and is probably buggy in other ways but I may still return to this, but not for a good long while I hope.